My research focus on solving realistic problem using machine learning and deep learning methods.
Inverse Force Design (current)
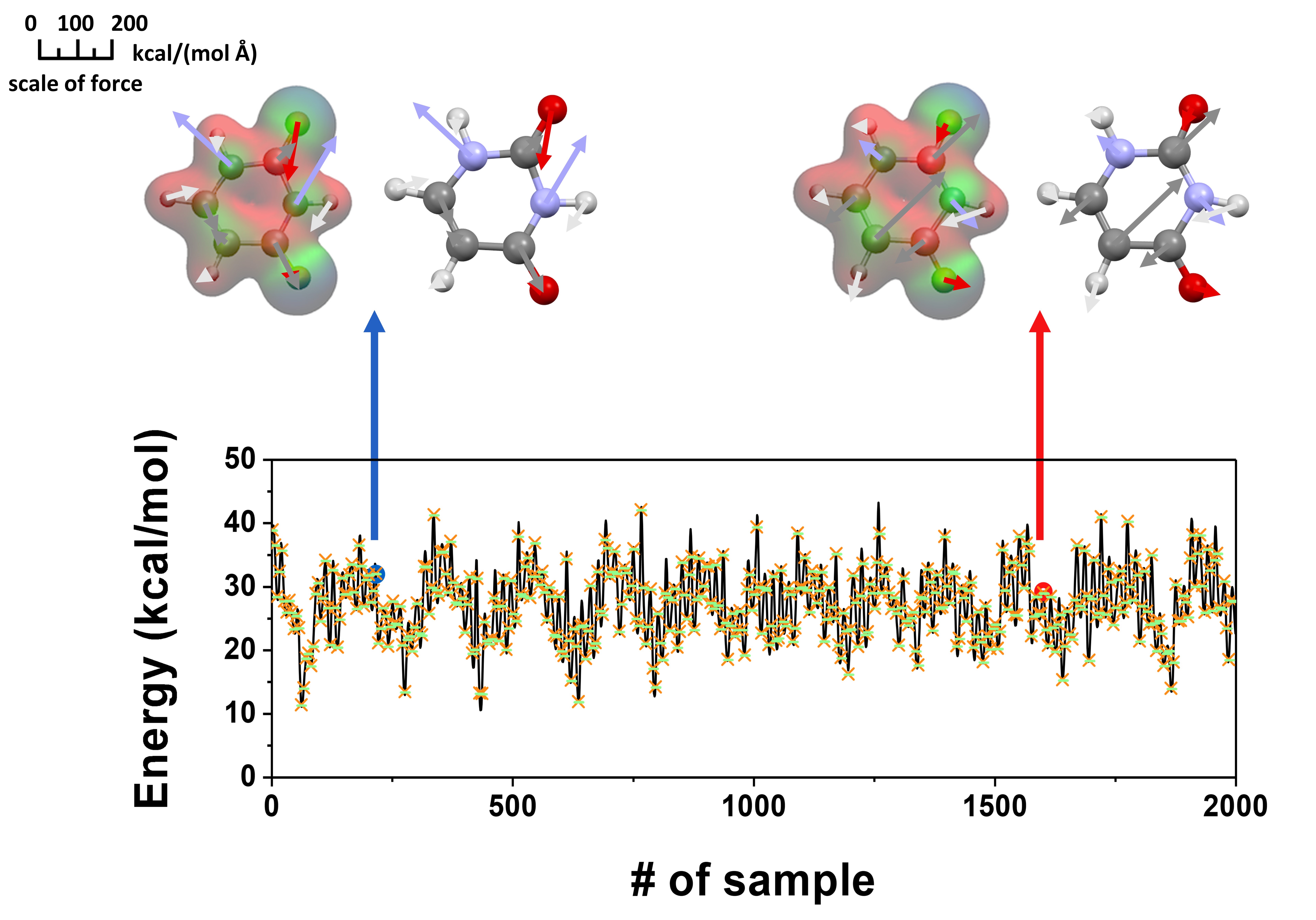
Quantum chemical methods reveal properties of a molecular system only after specifying the essential parameters of the constituent atomic nuclei and their three-dimensional (3D) coordinate positions. Inverse design, as its name suggests, inverts this paradigm by starting with the desired functionality and searching for an ideal molecular structure. Functionality need not necessarily map to one unique structure but to a distribution of probable structures. In this project, we focus on the inverse mapping of the Molecular Force Field. Given the desired force vectors on molecules such as H2CO, we are able to generate the structure
Efficient Machine Learning Force Field Emulation
Kernel ridge regression (KRR) that satisfies energy conservation is a popular approach for predicting forcefield and molecular potential, to overcome the computational bottleneck of molecular dynamics simulation. However, the computational complexity of KRR increases cubically as the product of the number of atoms and simulated configurations in the training sample, due to the inversion of a large covariance matrix, which limits its applications to the simulation of small molecules. Here, we introduce the atomized force field (AFF) model that requires much less computational costs to achieve the quantum-chemical level of accuracy for predicting atomic forces and potential energies. Through a data-driven partition on the covariance kernel matrix of the force field and an induced input estimation approach on potential energies, we dramatically reduce the computational complexity of the machine learning algorithm and maintain high accuracy in predictions.
